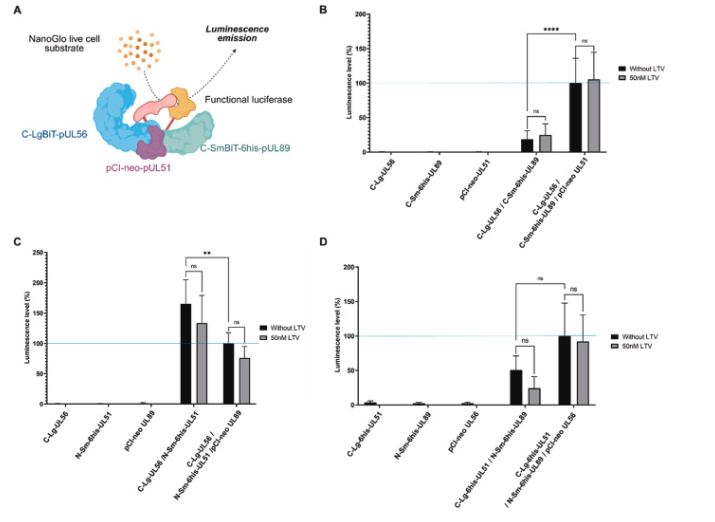
Cette publication est issue de la collaboration entre l’UMR 1092 – Anti-Infectieux : supports moléculaires des résistances et innovations thérapeutiques (RESINFIT) de Limoges et l’Institut Toulousain des Maladies Infectieuses et Inflammatoires (INFINITY) de Toulouse. Publiée dans la revue Antiviral Research, elle porte sur l’interaction des protéines du complexe terminase du CMVH en présence de létermovir et de mutations de résistance.